- Home
- Publications
- PAGES Magazine
- What Can APIs Do For You?
What can APIs do for you?
Gross W, Morrill C, Gille E and Shepherd E
Past Global Changes Magazine
30(1)
63
2022
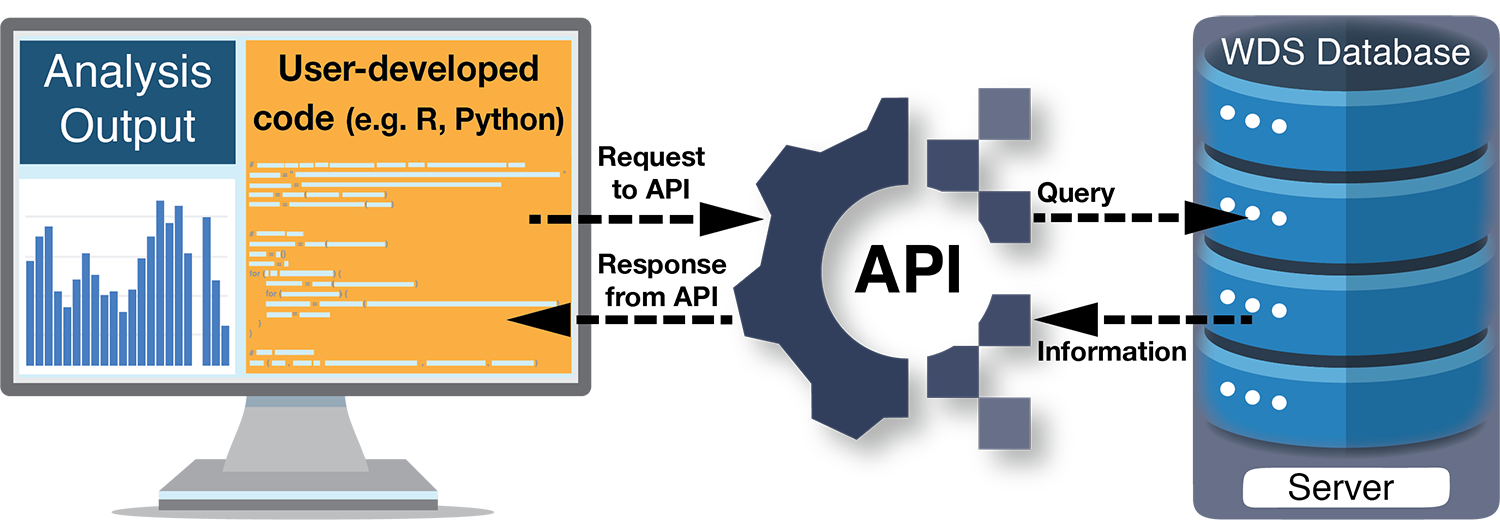
As the spatial and temporal resolution of scientific datasets and the culture of data sharing grow, more data on past global changes are now available than ever before. Efficiently discovering, downloading, and integrating data into analyses is critical for making full use of this surge of information. Researchers have long used Graphical User Interfaces1, or GUIs, to manually search and download data, but Application Programming Interfaces, or APIs, can handle these tasks in a more automated fashion. In fact, APIs are the behind-the-scenes conduit for many different types of information we use every day, from weather forecasts within smartphone apps to flight schedules aggregated on travel websites.
APIs are the technological backbone that let two computer programs communicate over the internet. Each API defines a set of rules that specify the parameters by which requests (or "calls") can be made, as well as the format of the computer-readable information that is provided in response. World Data System repositories such as the World Data Service for Paleoclimatology (WDS-Paleo)2, PANGAEA3, and Neotoma4 provide APIs, as do other data and information sources such as publishers (e.g. Springer, Wiley), publication databases (e.g. CrossRef, Web of Science), domain-specific databases (e.g. Global Biodiversity Information Facility), and analysis tools (e.g. ArcGIS).
A request to an API is usually written in the form of a specially-formatted web address, or Uniform Resource Locator (URL). Scientists can call an API by simply entering such a URL into their web browser or by incorporating calls to an API in data analysis code. These new capabilities open more automated ways of first finding and accessing information, and then integrating information, both from different sources, and with different analysis tools.
The ability to discover and download information programmatically via an API, as opposed to manually through a GUI, increases efficiency and diminishes the possibility for human error. For example, APIs make it easier to repeat a search (perhaps to find newly archived datasets or updates to existing datasets) or to gather information quickly for initial data exploration (perhaps to identify certain geographical areas or parameter types with sufficient amounts of data for an analysis). APIs can also make it more efficient to search multiple data providers. While requests must be structured to match the requirements of a specific API, they can be a useful way to locate data across several repositories. In fact, the federated data search provided by the WDS-Paleo API uses the Neotoma API to retrieve information about datasets.
API requests can be integrated with many programming languages (e.g. Python, R, MatLab), effectively creating a pipeline of data to tools for analysis. For example, functionality to access data from the PANGAEA, and Neotoma data repositories via their APIs exists in some Python5 and R6,7 packages (e.g. Goring et al. 2015) and the WDS-Paleo also provides example API requests in these languages8. Incorporating API requests into a scientific workflow promotes reproducibility and repeatability of research. Encoding all steps of the workflow from data discovery and download to processing and analysis provides complete documentation of the research methods, including the criteria used to select datasets. Some APIs also perform analysis directly: for example in the geospatial ecosystem, ArcGIS APIs9 and Open Geospatial Consortium APIs10 perform geospatial analysis, including image analysis and feature classification.
While APIs provide the ability to access formatted information directly from computer systems of authoritative sources, thereby streamlining data access and analysis, there are still some obstacles to integrating data from different repositories or sources seamlessly (e.g. EarthLife Consortium API11; Uhen et al. 2021). For example, interoperability and reusability of paleoenvironmental data also require enhanced common standards and workflows for metadata and data reporting (Bothe et al. 2021; Khider et al. 2019; Morrill et al. 2021; Williams et al. 2018). These improvements, in concert with technological advances such as APIs, will accelerate discovery from the many decades of data collection by the paleo community.
affiliationS
1National Centers for Environmental Information, National Oceanic and Atmospheric Administration, Boulder, CO, and Asheville, NC, USA
2Cooperative Institute for Research in Environmental Sciences, University of Colorado Boulder, CO, USA
3Riverside Technology, Inc, Fort Collins, CO, USA
contact
Wendy Gross: paleo noaa.gov
noaa.gov
references
Bothe O et al. (2021) PAGES Mag 29: 59
Goring S et al. (2015) Open Quat 1: Art 2
Khider D et al. (2019) Paleoceanogr Paleoclimatol 34: 1570-1596
Morrill C et al. (2021) Paleoceanogr Paleoclimatol 36: e2020PA004193
Uhen MD et al. (2021) Front Biogeogr 13: e50711
Williams J et al. (2018) PAGES Mag 26: 50-51
Links
1For example: ncei.noaa.gov/access/paleo-search, pangaea.de, apps.neotomadb.org/explorer
2ncei.noaa.gov/access/paleo-search/api
3ws.pangaea.de
4api.neotomadb.org/api-docs
5github.com/pangaea-data-publisher/pangaeapy
6docs.ropensci.org/pangaear
7cran.r-project.org/web/packages/neotoma
8ncei.noaa.gov/access/paleo-search/api#examples
9developers.arcgis.com/rest
10ogc.org/resource/demos-archive
11earthlifeconsortium.org