- Home
- Publications
- PAGES Magazine
- Sedimentary Ancient DNA (sedaDNA) As a New Paleo Proxy To Investigate Organismal Responses To Past Environmental Changes In Antarctica
Sedimentary ancient DNA (sedaDNA) as a new paleo proxy to investigate organismal responses to past environmental changes in Antarctica
Armbrecht L
Past Global Changes Magazine
30(2)
78-79
2022
The study of ancient DNA from sediments (sedaDNA) has great potential for paleoclimate research. Using less than a gram of sediment, this new technique allows ecosystem-wide assessments of Antarctic marine biodiversity over hundreds of thousands of years.
sedaDNA: A new paleo proxy
Marine sedimentary ancient DNA (sedaDNA) is DNA from dead organisms that have sunk from the ocean to the seafloor and been preserved there. Over time, layers of sedaDNA accumulate, forming a record of "who" has inhabited the ocean in the past. sedaDNA analysis is an interesting new paleo proxy because the genetic traces of organisms that do not fossilize can be detected, too (Capo and Monchamp et al. 2022). This means that sedaDNA allows us to study past marine biodiversity quite comprehensively across different levels of the food web, including bacterio- and phytoplankton, zooplankton, and potentially even fish, uncovering wide-scale community shifts as a response to past climatic change. Such knowledge is important, as it helps us to better predict the future of marine ecosystems with ongoing climate change and find management strategies to conserve them.
Antarctica: An important location for sedaDNA research
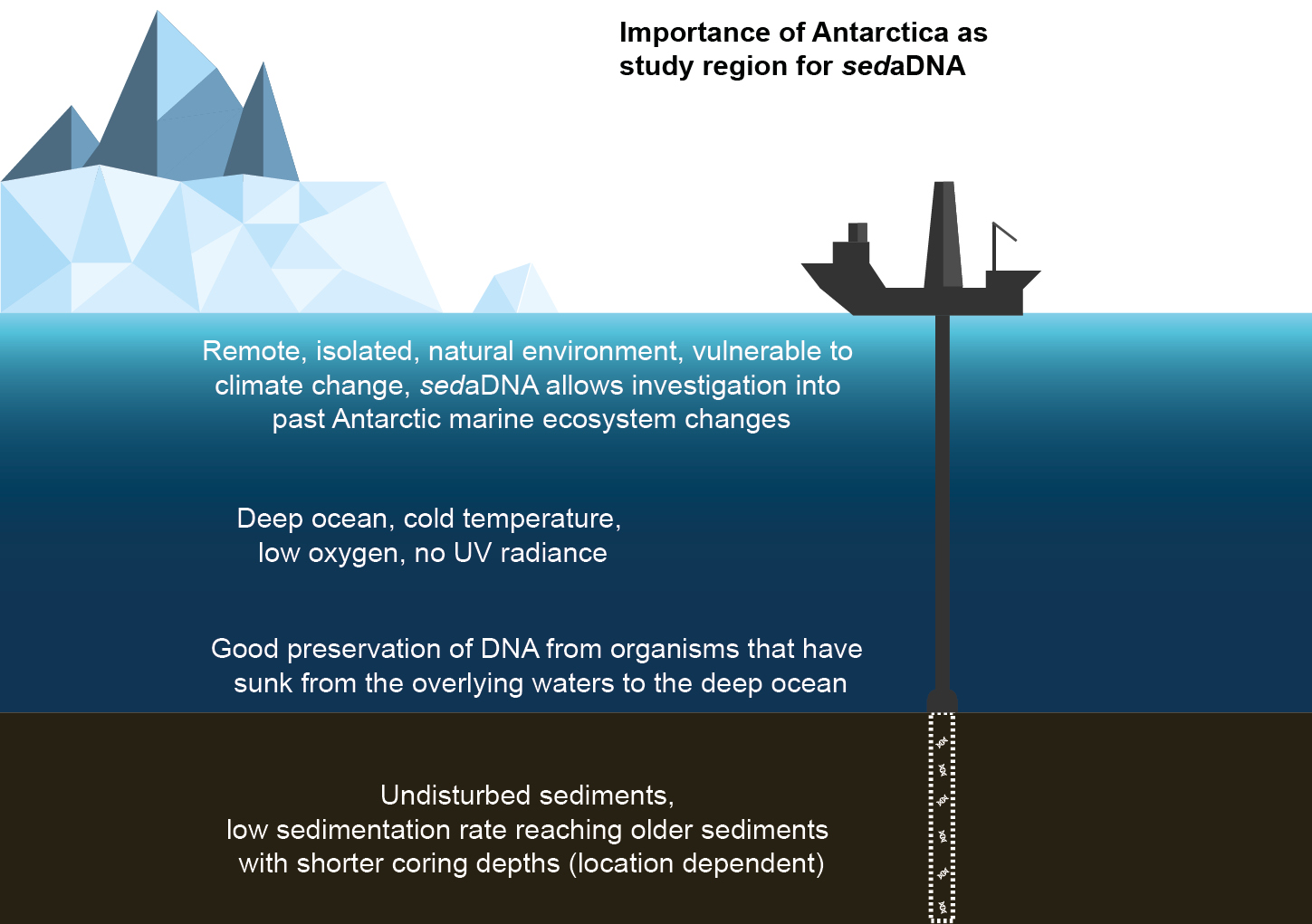
Polar deep-ocean environments are particularly suitable locations for sedaDNA research because they feature favorable conditions for sedaDNA preservation. These include constantly low temperatures and oxygen concentrations (~0°C, ~5 mL/L, respectively, noting that these values vary regionally; Bensi et al. 2022; Garcia et al. 2018; Meredith et al. 2008), and the absence of UV radiation (Karentz 1989). Antarctica and the Southern Ocean are remote and isolated, making them natural climate laboratories to study long-term global change (Barnes et al. 2006).
Sampling logistics in remote Antarctica are difficult, and for sediment studies in particular, large research vessels or platforms are required to have the capacity to drill into the deep seafloor, sometimes several thousands of meters below the ocean surface (Fig. 1). The most suitable coring system to acquire sediments for sedaDNA analysis is piston coring, which "punches a hole" into the seafloor (rather than using active drilling) and thus recovers undisturbed sediments (Armbrecht et al. 2019). The reliance on piston coring means that sedaDNA analyses are restricted to relatively soft sediments, usually found in the upper sediment layers. However, this is not necessarily a limitation – the recovery of sediments of up to ~490 m below the seafloor has been achieved using piston coring (Tada et al. 2015), which, in many Southern Ocean regions, can reach sediments of ages that are far beyond the timescales that allow for detection of ancient DNA.
Deep Southern Ocean sediments have relatively low sedimentation rates compared to coastal areas. For example, in >3,000 m water depth in the Scotia Sea, sedimentation rates have been determined at ~10–40 cm per 1,000 years (in the upper ~430 m; Weber et al. 2021). Thus, even relatively shallow coring can provide access to sediments of considerable age, allowing sedaDNA investigations into changes in marine food web structures over multiple glacial–interglacial cycles.
Consequently, the limitation on how far back in time ancient DNA analyses can be applied to deep ocean sediments remains not a coring capacity question, but rather one of maximum age of sedaDNA preservation. It is expected that ancient DNA can be preserved for up to ~1 million years under the right conditions (although reports exist of non-replicated/authenticated ancient DNA from bacteria reaching several millions of years; Willerslev and Cooper 2005, and references therein). Until recently, the oldest authenticated sedaDNA had been from terrestrial systems (cave sediments) that were ~400,000 years old (Willerslev et al. 2003). In the Arctic environment, eukaryote sedaDNA has been found in up to 140,000-year-old sediments (Pawłowska et al. 2020). In the Antarctic, marine eukaryote sedaDNA has recently been found in ~1 million-year-old sediments in the Scotia Sea (Armbrecht et al. 2022).
Current applications of sedaDNA research in the Antarctic
Contamination-free sampling techniques are starting to be more commonly used on board research vessels, and sedaDNA research is becoming more frequently incorporated into Antarctic science. For example, in 2019, extensive sedaDNA sampling was undertaken during IODP Exp. 382 "Iceberg Alley and Subantarctic Ice and Ocean Dynamics", using some of the most stringent anti-contamination procedures to date (Weber et al. 2021). In addition to clean sampling (via the use of sterilized core-cutting and sampling equipment), the use of non-toxic chemical tracers to determine potential contamination of the core liners (which can occur during the hydraulically driven piston coring process) was benchmarked in the context of sedaDNA research during this expedition (Weber et al. 2021). Previously, this technique had been routinely used by geomicrobiologists when collecting deep biosphere samples for the study of actively living microbial communities, where contamination by modern microbes is of paramount concern (Sylvan et al. 2021).
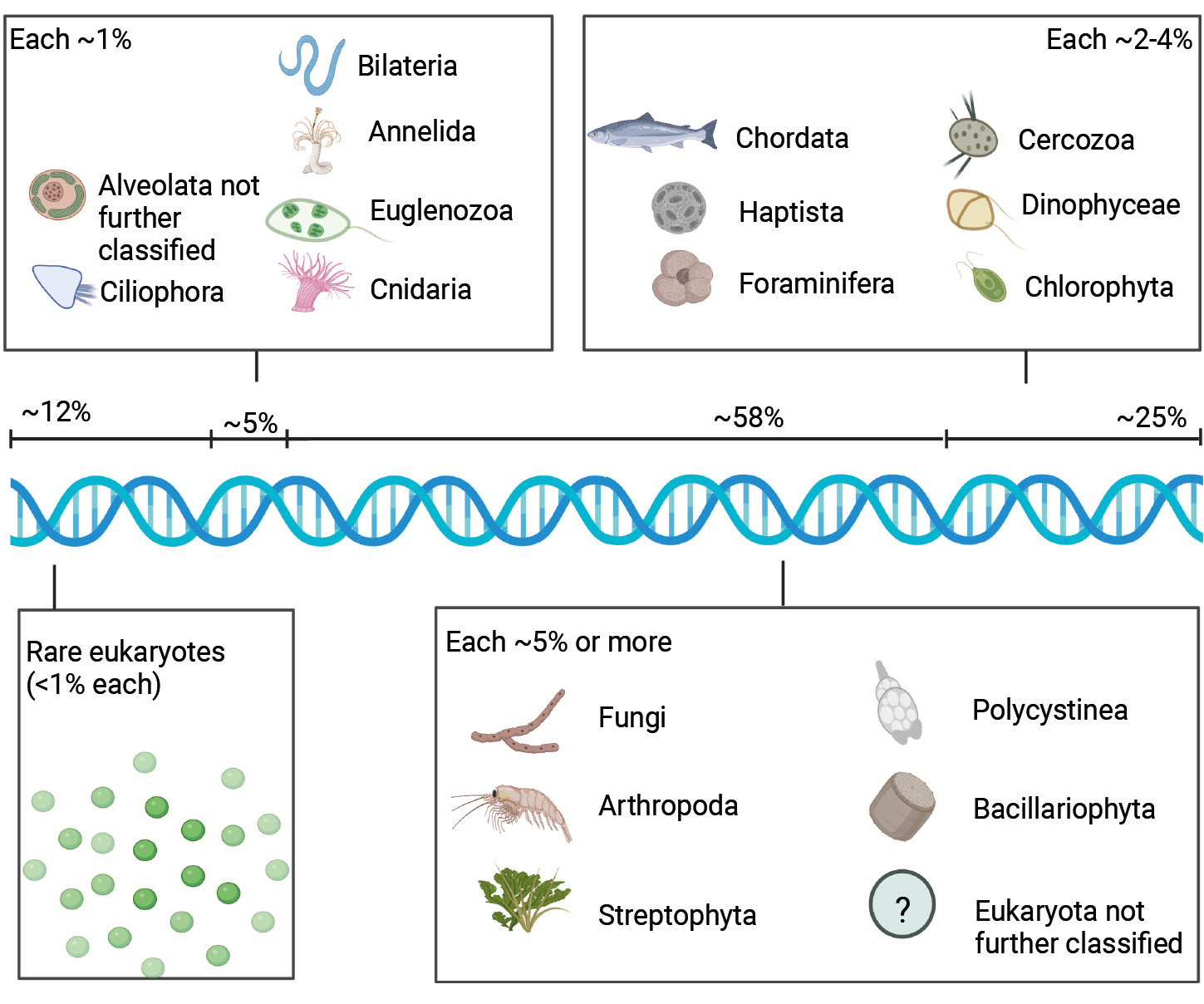
The sedaDNA analyses of IODP Exp. 382 samples aimed at the detection of different taxonomic marker genes (genes that are variable enough in their sequence so species-specific determination is possible) to identify marine eukaryotes, including the small and large subunit ribosomal RNA genes (SSU, LSU) and the Photosystem II manganese-stabilizing polypeptide gene (psbO, which only occurs in photosynthesizing organisms; Pierella Karlusich et al. 2022). Both fossilizing and non-fossilizing eukaryotes were detected, including diatoms and chlorophytes (back to ~540,000 years), as well as a range of other eukaryote groups (Fig. 2). This shows that research into many groups of organisms over hundreds of thousands of years using sedaDNA analyses is feasible, and especially so in Antarctica and the Southern Ocean.
Outlook for sedaDNA research in Antarctica
The potential of sedaDNA as a paleo proxy is in (1) its ability to complement the fossil record through the detection of ancient DNA from organisms that don't normally fossilize or otherwise allow for reconstructions of the marine food web, and (2) the possibility to study not only biologic composition of various sites ("who was there") but also the activity and function of organisms that lived there in the past ("what were they doing"). In the Antarctic sea-ice environment, such organisms of interest may, for example, include various fragile diatoms that could be useful as sea-ice proxies (e.g. highly branched isoprenoid producing species; Zimmermann et al. 2020) or other primary producers, such as chlorophytes and non-cyst forming/fragile dinoflagellates (De Schepper et al. 2019). Antarctic krill are also highly abundant in sea-ice environments, though they are currently experiencing hardship due to ocean acidification, warming, and overfishing (Flores et al. 2012). sedaDNA analysis makes it possible to track the presence and dynamics of these important Antarctic species over geological timescales.
 |
|
Figure 2: Overview and proportions of eukaryote groups that can be detected using sedaDNA in the Antarctic region. Approximate proportions (percentage of eukaryote groups out of total eukaryotes) are based on Armbrecht et al. (2022). Figure created with BioRender.com (note that icon selection depended on availability in BioRender and may not necessarily depict Antarctic species). |
Despite significant progress in sedaDNA research during recent years, the discipline is still in its infancy, with some baseline research questions needing to be addressed. For example, preservation biases are important to consider when interpreting sedaDNA data, yet little is known about such biases. It has been shown that sedaDNA degradation correlates with organic matter degradation (Armbrecht et al. 2022), but how well the DNA of certain species is preserved compared to that of others, and how far DNA can be transported with deep ocean currents, is currently unknown but would dramatically improve the accuracy of sedaDNA-derived ecosystem reconstructions.
sedaDNA is only preserved in trace amounts in the deep seafloor, and this scarcity makes it difficult to investigate rare species, which might sometimes be the most suitable indicators for specific environmental conditions. To overcome the hurdles of rare sequence detection in marine sedaDNA samples, high sequencing depths (acquiring many millions of reads) per sample is recommended and is becoming more affordable with the availability of today's next generation sequencing platforms. RNA-based hybridization capture techniques that enrich specific (e.g. rare) target sequences (Horn 2012) might further allow for more detailed investigations into higher-trophic-level organisms such as fish.
In summary, recent improvements in sedaDNA acquisition and analysis techniques in combination with sediment samples from locations characterized by ideal sedaDNA preservation conditions, such as those in polar ecosystems, make the application of this new proxy particularly promising for Antarctic paleo research, and open new doors to food-web-wide reconstructions over hundreds of thousands of years in this vulnerable, remote region. The depth and detail of the picture that sedaDNA can give us of past marine life is only just beginning to be explored.
affiliation
Institute for Marine and Antarctic Studies (IMAS), University of Tasmania, Hobart, Australia
contact
Linda Armbrecht: linda.armbrecht utas.edu.au
utas.edu.au
references
Armbrecht L et al. (2019) Earth-Sci Rev 196: 102887
Armbrecht L et al. (2022) Nat Commun 13: 5787
Barnes DKA et al. (2006) Glob Ecol Biogeogr 15: 121-142
Bensi M et al. (2022) Earth Syst Sci Data 14: 65-78
Capo E, Monchamp ME et al. (2022) Environ Microbiol 24: 2201–2209
De Schepper S et al. (2019) ISME J 13: 2566-2577
Flores H et al. (2012) Mar Ecol Prog Ser 458: 1-19
Horn S (2012) In: Shapiro B, Hofreiter M (Eds) Ancient DNA. Springer, 177-188
Karentz D (1989) Antarct J US 24: 175-176
Meredith MP et al. (2008) J Clim 21: 3327-3343
Pawłowska J et al. (2020) Sci Rep 10: 15102
Pierella Karlusich JJ et al. (2022) Mol Ecol Res doi: 10.1111/1755-0998.13592
Sylvan JB et al. (2021) Technical Note 4. International Ocean Discovery Program, 16 pp
Tada R et al. (2015) In: Tada et al. (Eds) Proc IODP 346.Integrated Ocean Drilling Program, 1-61
Weber ME et al. (2021) Proc IODP 382. International Ocean Discovery Program), 309 pp
Willerslev E, Cooper A (2005) Proc Royal Soc B 272: 3-16