- Home
- Publications
- PAGES Magazine
- The Living Record: Considerations For Future Biological Studies of Ice Cores
The living record: Considerations for future biological studies of ice cores
Willis MC, Chellman N & Smith HJ
Past Global Changes Magazine
30(2)
110-111
2022
Madelyne C. Willis1,2, N. Chellman3 and H.J. Smith2,4
This article highlights the state of knowledge of glacial microorganisms, focusing on englacial habitats, challenges associated with studying cells in these environments, and considerations for future ice-core projects seeking to advance biological studies as part of their scientific objectives.
Once thought to be inhospitable to life, glaciers and ice sheets are now considered microbially dominated biomes. Anesio et al. (2017) estimated there may be on the order of 1029 cells in all of Earth's glaciers and ice sheets, on the same order of magnitude as the reported total cell abundance for all aquatic systems on Earth (1.2 x 1029 cells; Whitman et al. 1998). Originally assumed to be preserved in a dormant state, studies over the past 20 years have demonstrated many of these cells are likely viable, and their presence and function have profound implications for a wide range of scientific fields including paleoclimatology, bioprospecting, and exobiology (D'Andrilli et al. 2017; Balcazar et al. 2015; Tung et al. 2005). Despite this shift to a perception of glaciers as habitable, methodological challenges and the fact that biological studies are often secondary to other scientific goals on deep ice coring projects have limited the study of microorganisms in englacial ice. Looking forward, recent advancements in lab- and field-based methods have created new opportunities for investigating life in these unique ecosystems.
Implications of ice as a habitable space
Glaciers and ice sheets contain liquid water features which may be habitable for microorganisms throughout all three glacial zones (supraglacial, englacial and subglacial) (Boetius et al. 2015). Investigations of glacier microbial communities have focused primarily on the relatively dynamic supraglacial and subglacial zones, emphasizing surface features such as ephemeral meltwater streams and ponds, and cryoconites (depressions in the surface filled with dust and liquid water; Cook et al. 2015), and subglacial hydrological systems (Mikucki et al. 2016; Walcott et al. p. 114). Much less is known about the biology of englacial ecosystems, despite these environments comprising the bulk of glacier ice mass (Boetius et al. 2015).
Within the englacial environment, habitable spaces may be found on the micron scale in water-filled pore spaces between ice crystals and in thin layers of liquid water surrounding dust particles trapped within ice (Tung et al. 2005). While a lack of energy sources and nutrients in these microhabitats may inhibit optimal growth, it is widely accepted that under these conditions microbes can maintain the low levels of activity needed to support basic housekeeping functions (Dieser et al. 2013). These functions, for example DNA repair, allow the cell to remain viable and may result in the uptake or production of some greenhouse gases (Fig. 1). Over geologic timescales, the activity required for cellular maintenance may be adequate to offset ice-core gas records by producing anomalous, non-atmospheric signals of gases e.g. nitrous oxide, methane, and carbon monoxide (Miteva et al. 2016; Fain et al. 2022; Banerjee et al. p. 104). At present, our understanding of in-situ microbial activity within glacier ice is limited to either theoretical (Tung et al. 2005), or in-vitro laboratory studies (Dieser et al. 2013); there has been no direct measure of microbial activity within deep glacial ice. Studies providing empirical evidence of microbial activity or quiescence would facilitate more robust paleoclimatic reconstructions, and understanding the resilience of these organisms may inform our search for life on Mars or other planetary environments containing water ice.
Challenges and considerations
The gap in knowledge regarding in-situ biological activity is largely due to the difficulty of performing biological measurements on ice-core samples. The primary hurdle for most studies is the inherently low biomass within glacier ice. Although cell concentrations as high as 106 cells/mL have been reported (Miteva et al. 2016), these high numbers correlate with high dust concentrations and, in general, englacial cell numbers tend to be much lower: between 101 and 104 cells/mL (Santibáñez et al. 2018).
The challenge of low biomass is exacerbated by limited sample volumes available from deep ice-core projects, contamination, and insufficient sensitivity of analytical methods. Core fracturing is a source of contamination that can easily be introduced during ice-core breaking and inconsistent temperature storage. Contamination from mechanical drilling practices that use hydrocarbon-based drilling fluids is of particular concern, as these fluids can contaminate both cores and the subglacial environment. Existing ice-core decontamination protocols are effective but can result in appreciable sample loss. Once samples have been transported to the laboratory and decontaminated, many traditional microbiological approaches lack the sensitivity required for low biomass englacial ice. Depending on final cell concentrations, relatively large sample volumes (5–500 mL meltwater) are often required for these approaches.
Recent developments
Fortunately, recent advancements in drilling systems, microbial analytical methods, and in-situ technology make this an exciting moment for probing questions about microbiology in ice. Hot-water drilling and air-reverse circulation are alternatives to mechanical drilling with organic fluids and have been demonstrated to be effective and to limit contamination (Talalay and Hong 2021). In addition, engineering solutions which prevent vertical and diagonal fracturing of cores during drilling processes preserve more core sections suitable for microbial analysis (Talalay and Hong 2021). Use of a replicate ice-coring system can provide additional sample volume at depths with high community demand for core sections by drilling replicate cores slightly deviated from the original borehole. Use of these systems could provide the sample volume required for microbial analyses.
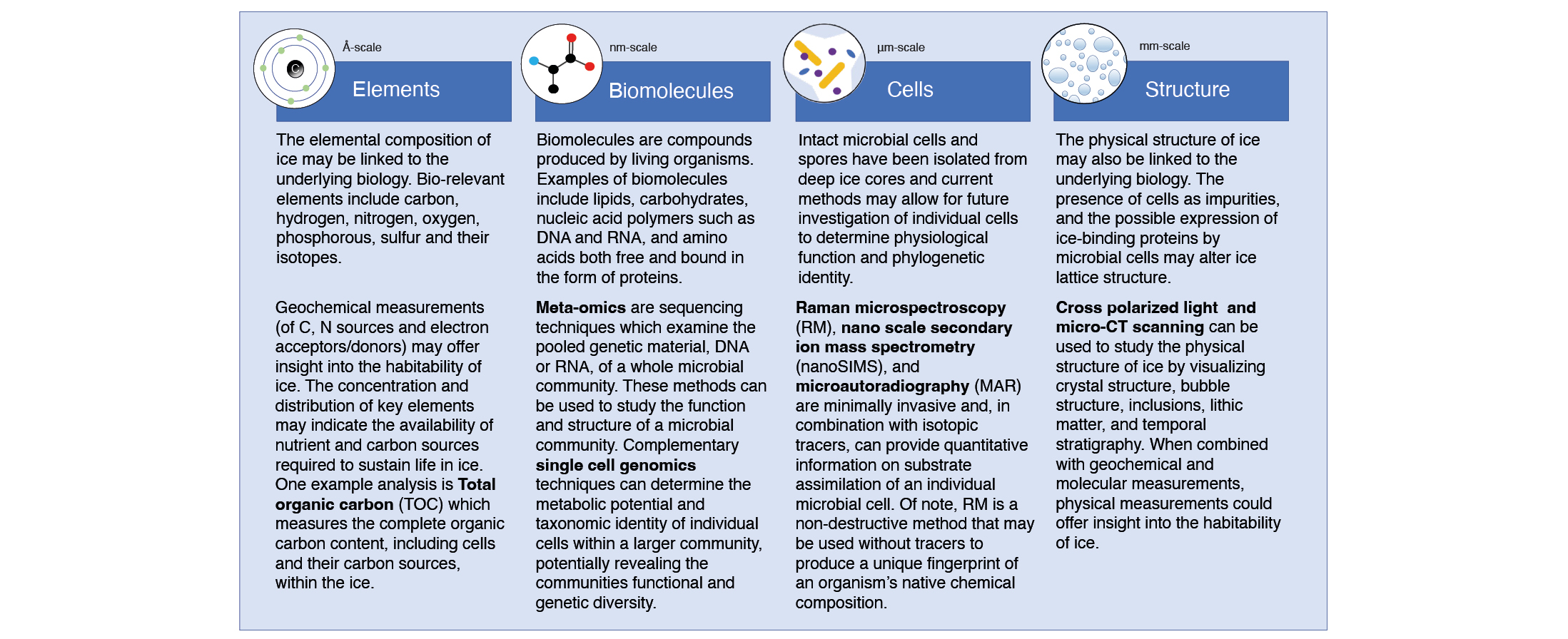
In the lab, continuous flow analysis provides detailed temporal resolution of decontaminated ice (Santibáñez et al. 2018), which is particularly useful for biological applications to monitor contamination (e.g. the detection of drilling fluids or other anomalies). Innovative and highly sensitive analytical techniques, such as nanoSIMS, stable isotope probing, and other next-generation physiology measurements can reveal cellular function on the single-cell level (Fig. 2). Excitingly for ice-core studies, many of these next-generation approaches are also nondestructive, which enables crucial downstream analyses of individual cells such as cultivation, sequencing, and "omics" approaches. These methods have been demonstrated for studies of microbial diversity and physiology in a variety of low-biomass natural samples; however, they have yet to be applied to studies of deep ice cores. Hatzenpichler et al. (2020) provide a full review of next-generation physiology techniques.
Field-deployable technologies are complementary to lab-based methods and are capable of detecting cells or biorelevant compounds within the solid ice matrix (Eshelman et al. 2019). Cells and compounds that may become too dilute once melted (ex: 102–104 cells/mL), can be concentrated at detectable levels (106–108 cells/mL) within the grain boundaries of solid ice (Mader et al. 2006). Since most biological measurements traditionally require samples to be melted before analysis, the development of non-destructive technologies could result in new approaches to studying englaciated life in situ. Additionally, the incorporation of these technologies into the drilling process creates the potential for real-time data collection within the ice borehole. This could provide a means to detect areas of interest based on organic or microbial content during the drilling process and allow for data-driven decision-making during ice-core collection, for instance in determining the depths of interest for replicate coring.
Conclusion
Ice-core research has traditionally focused on reconstructing Earth's climate and environmental history using measurements of stable water isotopes, gases, and other inorganic compounds preserved within the ice. However, we now have the capability to better understand the abundance and function of microbial communities in ice. These organisms may have a profound impact on paleoclimatic records preserved in ice chemistry, may be used as additional indicators of past depositional events related to climate, and may serve as proxies for life in extraterrestrial water ice elsewhere in our solar system. If considerations for biological measurements are taken into account early in planning future drilling projects, there will be greater opportunities to discover the englacial microbiome.
Acknowledgements
The authors would like to thank Foreman Lab Group members for discussions about life in ice and comments that improved the manuscript. M. Willis, and H. Smith are supported by NSF Antarctic Research (2037963).
affiliationS
1Department of Land Resources and Environmental Science, Montana State University, Bozeman, USA
2Center for Biofilm Engineering, Montana State University, Bozeman, USA
3Division of Hydrologic Sciences, Desert Research Institute, Reno, NV, USA
4Department of Microbiology and Cell Biology, Montana State University, Bozeman, USA
contact
Madelyne Willis: madie.willis gmail.com
gmail.com
references
Anesio AM et al. (2017) NPJ Biofilms Microbiomes 3: 10
Balcazar W et al. (2015) Microbiol Res 177: 1-7
Boetius A et al. (2015) Nat Rev Microbiol 11: 677-690
Cook J et al. (2015) Prog Phys Geogr 40: 66-111
D'Andrilli J et al. (2017) Geochem Perspec Lett 4: 29-34
Dieser M et al. (2013) Appl Environ Microbiol 24: 7662-7668
Eshelman EJ et al. (2019) Astrobiology 19: 771-784
Faïn X et al. (2022) Clim Past 18: 631-647
Hatzenpichler R et al. (2020) Nat Rev Microbiol 18: 241-256
Mader HM et al. (2006) Geology 34: 169-172
Mikucki JA et al. (2016) Philos Trans R Soc A 374: 20140290
Miteva V et al. (2016) Geomicrobiol J 33: 647-660
Santibáñez PA et al. (2018) Glob Chang Biol 24: 2182-2197
Talalay PG, Hong J (2021) Ann Glaciol 62: 143-156
Tung HC et al. (2005) Proc Natl Acad Sci USA 102: 18,292-18,296
Whitman WB et al. (1998) Proc Natl Acad Sci USA 95: 6578–6583