- Home
- Publications
- PAGES Magazine
- Next Generation Sequencing, Phylogeography, and Paleoecology
Next generation sequencing, phylogeography, and paleoecology
Ficetola GF & Taberlet P
Past Global Changes Magazine
28(1)
10-11
2020
Gentile F. Ficetola1,2 and Pierre Taberlet2,3
Genome skimming and environmental DNA extracted from lake sediments are increasingly important to measure genetic diversity, and to understand how environmental changes affected species distribution through time. Here we describe how genome skimming on the current geographic distribution of species can be combined with the analysis of lake sediments, to perform unprecedented tests of phylogeographic hypotheses, and reconstruct past refugia and colonization routes.
DNA sequencing has greatly improved over the past 15 years, and next generation sequencing is transforming DNA analysis (van Dijk et al. 2014). Today, the most powerful sequencer, the NovaSeq 6000 (Illumina), is able to produce up to 40 billions of sequence reads of 250 nucleotides in a single experiment. Such sequencing power induces a complete change in assessing genetic diversity of wild species. It is now affordable to analyze a significant portion of the genomes instead of looking only at a few target regions. Here, we will focus on two approaches linked to next generation sequencing: genome skimming and environmental DNA analysis; both can greatly improve our understanding of how environmental changes have impacted the genetic diversity of species.
Genome skimming (Coissac et al. 2016; Dodsworth 2015; Straub et al. 2012) corresponds to the sequencing of random DNA fragments of the genome, at a sequencing depth that does not allow for the assembly of the whole nuclear genome, but that allows for the assembly of all repetitive DNA such as, for plants, the chloroplast genome, the nuclear ribosomal DNA, and possibly the mitochondrial genome. Genome skimming has been proposed as an extension of the current barcoding approach (Coissac et al. 2016), as it provides much more information for DNA-based species identification and for phylogenies (e.g. Malé et al. 2014).
The analysis of environmental DNA extracted from lake sediments allows us to reconstruct the variation of species distribution over the last few millennia, and is becoming a widely used approach for assessing past communities, either targeting a single species, or dealing with all species from a taxonomic group such as plants or mammals (review in Bálint et al. 2018; see also Giguet-Covex et al. 2014; Pansu et al. 2015). Although most of the studies performed so far focused on individual species or on higher-level taxonomic entities, lake sediments could also be a source of information at the intraspecific level, provided that the appropriate DNA markers are available.
Combining genome skimming and environmental DNA
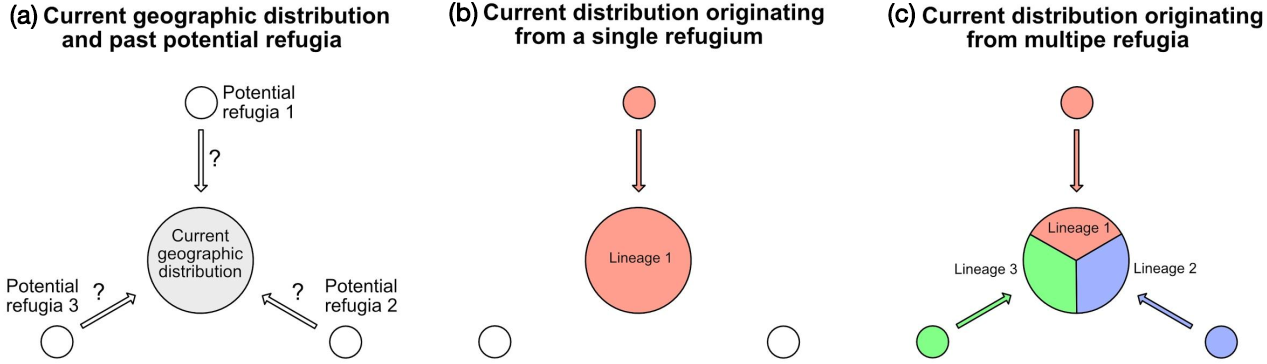
By combining genome skimming on the current geographic distribution of a target species, and the analysis of lake sediments, it is now possible to test intraspecific phylogeographic hypotheses, i.e. to find past refugia and colonization routes. Figure 1 illustrates this approach, which has not yet been implemented but has a great potential for understanding past distributions of different lineages within a species.
 |
|
Figure 1: Hypothesis testing, based on phylogeographic data and lake sediment analysis in different potential refugia (see text for details). |
The first step must start with a phylogeographic study of the considered species. This consists of collecting representative samples over the whole current distribution (Fig. 1a). After DNA extraction, these samples are sequenced on a next generation platform, using a genome skimming approach, to produce about one gigabase of sequence, e.g. about seven million sequence reads, each 150 base pairs long. Based on these sequences, for each sample, the whole chloroplast DNA and the whole nuclear ribosomal DNA can be assembled. The comparison of these sequences among all samples reveals the phylogeographic structure of the species, showing the geographic distribution(s) of either a single homogeneous lineage (Fig. 1b), or different lineages (Fig. 1c).
The second step is the identification of genetic markers that unambiguously characterize each lineage, with the goal of being able to recover these markers from lake sediments. This requires choosing markers that are as short as possible, with conserved flanking regions where PCR (polymerase chain reaction) primers, i.e. short DNA sequences that are necessary for DNA amplification, can be anchored. As genome skimming provides the sequences of the whole chloroplast DNA and of the ribosomal DNA, it is quite easy to find such short diagnostic markers for each lineage.
The third step consists of collecting lake sediments, within both the current geographic distribution and potential refugia. The different cores must be precisely dated, and DNA extraction is carried out from many core slices. The objective is to test for the presence or absence of the different lineages at different times and locations. This presence is determined using the DNA markers designed in step two. These markers are amplified via PCR and sequenced on next generation sequencers. The results should allow us to reconstruct the history of each lineage very precisely, including past presence in areas where it later disappeared, and colonization routes towards the current geographic distribution.
Perspectives
All the technologies and methodologies for carrying out the above approach have already been available for more than five years. Surprisingly, not a single paper has taken the opportunity to combine them for resolving important controversies concerning potential refugia, postglacial colonization routes, and the evolution of intraspecific genetic diversity. For example, the re-establishment of flora in Scandinavia is controversial, opposing the "tabula rasa" versus the "nunatak" hypotheses (Brochmann et al. 2003). The "tabula rasa" hypothesis suggests that after the last glacial maximum, Scandinavia was only recolonized from the South. Alternatively, the "nunatak" hypothesis opens the possibility for a recolonization from non-glaciated cryptic refugia in Scandinavia such as Andøya Island (Parducci et al. 2012). The recolonization of Scandinavia could be tested by collecting lake sediments in the plain south of the ice sheet during the last glacial maximum. If for many species the same lineages are found in Scandinavia and in these plains where they are currently absent, this will be in favor of the "tabula rasa" hypothesis. On the contrary, if the lineages in these plains about 20 thousand years ago are different from the ones found in Scandinavia, this will support the possibility of northern cryptic refugia.
Climatic oscillations occurring since the Pleistocene have shaped the present day biodiversity of plant and animal species; yet, human-driven global changes are leading to unprecedented changes of species distributions and diversity. Understanding the processes that have determined the present-day genetic diversity can also help us understand the biotic response to ongoing environmental changes, and identify appropriate management strategies.
affiliations
1Department of Environmental Science and Policy, Università degli Studi di Milano, Italy 2Laboratoire d'Ecologie Alpine (LECA), CNRS, Université Grenoble Alpes, France 3The Arctic University of Norway (UiT), Tromsø Museum, Norway
contact
Gentile Ficetola: francesco.ficetola gmail.com
gmail.com
references
Bálint M et al. (2018) Trends Ecol Evol 33: 945-957
Brochmann C et al. (2003) Taxon 52: 417-450
Coissac E et al. (2016) Mol Ecol 25: 1423-1428
Dodsworth S (2015) Trends Plant Sci 20: 525-527
Giguet-Covex C et al. (2014) Nat Commun 5: 3211
Malé P-JG et al. (2014) Mol Ecol Resour 14: 966-975
Pansu J et al. (2015) Mol Ecol 24: 1485-1498
Parducci L et al. (2012) Science 335: 1083-1086